Compound Fetching from ChEMBL
ChEMBL (opens in a new tab) is a curcurated chemical database which provides bioactive molecules withdrugs like property and also provides chemical, bioactive & genomic data to aid the translation of genome information into effective new drugs.
ChEMBL database
RESTful Web Services

In this image every oval is a resource on the chembl database like when we search chemical id the image resource show images related to that chemical id, Activity show s the activity related to the molecular bioactivity.
Compound Activity Measures
IC50 measure
*it is a inhibitory potency of the substance in 50% of the population, and the biological target could be enzyme, cell receptor or microorganism, IC50 values are expressed in molar concentration.*
- Half maximal inhibitory concentration
- indicates how much of a paticular durg or other substance is needed to inhib a given biological process by half
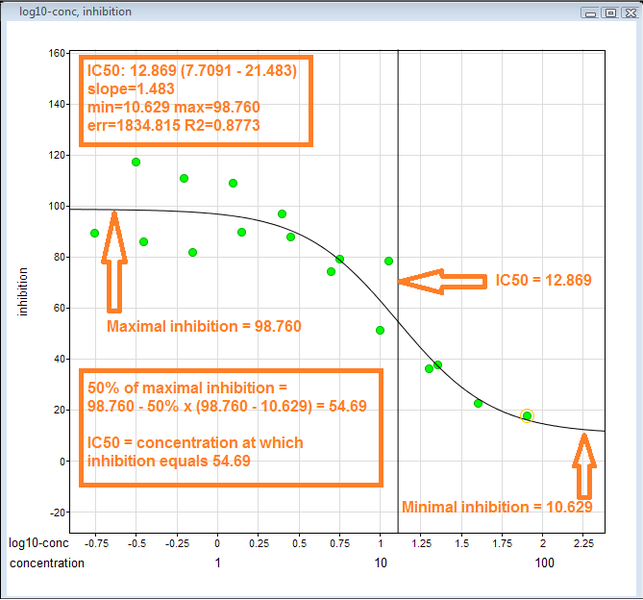
This is the visual representation of how to derive ic50 value 1. Arrange inhibitory data on y-axis and log(conc) on x-axis. 2. Identify max and min inhibition. 3. The IC50 is the con at which the curve passes through the 50% inhibition level
pIC50 value
-
To facilate the comparision of IC50 values, which have a large value range and given in different units (M,nM,...), often pIC50 values are used.
-
The pIC50 is the negative log of the IC50 Value when converted to molar units :
other activity measures:
Besides, IC50 and pIC50, other bioactivity measures are used, such as the equlibrium constant KI and half-maximal effective concentration EC50.
Pratical
In this pratical shown in course we are going to download all the molecules that have been tested on our target of intrest epidermal growth factor receptor (EGDR) Kinase
Connect to ChEMBL database
First all the libraries are imported, i have implementd this code on google colab so there may be some extra lines of code 😎.
# to download rdkit,chembl_webresource_client on the system
!pip install rdkit
!pip install chembl_webresource_client
import math
from pathlib import Path
from zipfile import ZipFile
from tempfile import TemporaryDirectory
import numpy as np
import pandas as pd
from rdkit.Chem import PandasTools
from chembl_webresource_client.new_client import new_client
from tqdm.auto import tqdm